Research Overview
The BMDS Lab conducts research across a broad spectrum of areas in artificial intelligence, machine learning, data science, and their applications in biomedical and health-related domains.
Research Areas
- Artificial Intelligence (AI): Neuro-inspired algorithms; cognitive modeling based on perception-action cycle, memory, attention, intelligence, and language.
- Machine Learning (ML): Deep generative models; unification of neural networks and probabilistic graphical models; deep network learning; large-scale sparse matrix factorization; sparse representations; disentangled representation learning; generalized and regularized linear models; kernel/nonlinear methods; deep feature selection; Bayesian learning and inference.
- Data Science: Theories and applications of big data analytics; structured and unstructured data modelling; heterogeneous data integration for knowledge discovery and actionable decision making; multi-view matrix factorization; multi-modal deep learning models; network science; data visualization; knowledge representation and knowledge learning from complex data.
- Bioinformatics: Annotation of non-coding regions of the human genome; biomarker identification using next-generation/deep sequence data; systems biology; high-dimensional biological data analysis.
- Chemoinformatics: Molecular representation; drug design and repurposing; RNA and protein structure prediction; biomolecular interaction prediction.
- Health Informatics: Cancer research; personalized disease diagnosis and treatment; biomedical image processing; intelligent and integrative health-informatics systems.
- Optimization: Fast numerical optimization methods for large-scale ML and data mining models; multi-objective optimization in ML and design tasks
- Computational Intelligence: Bio-inspired computation; quantum computing for ML.
AI for Drug Design
Drug discovery is the process of finding useful compounds that have a therapeutic effect towards a health condition. It aims to select molecules that satisfy multiple pharmacodynamic properties (e.g., specific binding to a target) and pharmacokinetic properties (e.g., absorption, distribution, metabolism, excretion, and toxicity), while ensuring efficacy and safety. This process is expensive, time-consuming, and with a high degree of uncertainty regarding the likelihood of a drug's success. The chance of identifying one molecule that fulfills all requirements, in a massive library and following sequential screening steps, is rather rare. The search space is huge up to 10⁶⁰ and brutally enumerating each molecule is computational infeasible. On average, it takes billions of dollars and more than 10 years to successfully have a drug reach the approval stage. The recent rise of AI techniques has provided a promising opportunity to revolutionise the drug discovery process.
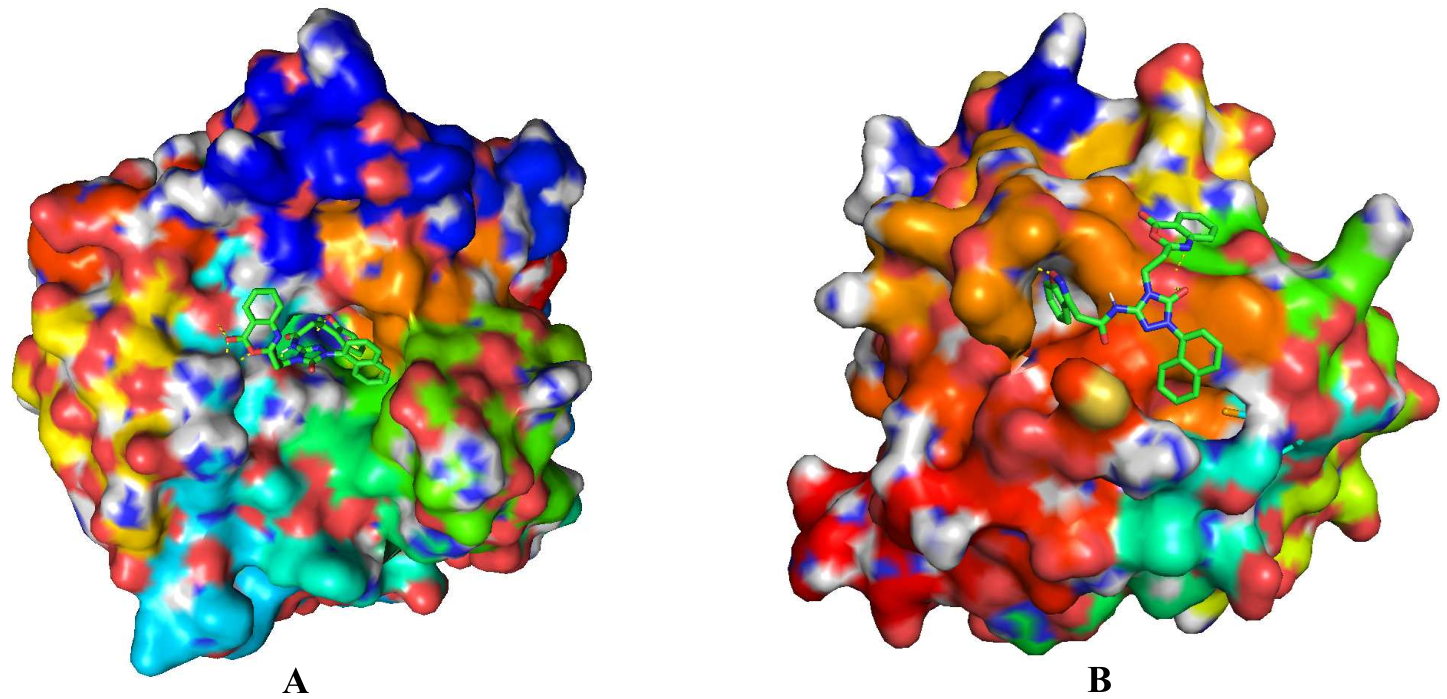
At the BMDS Lab, new AI approaches have been developed to design drug candidates that simultaneously satisfy multiple properties. For example, the deep evolutionary learning (DEL) approach published in [3,4] combines the strengths of deep generative models and multi-objective evolutionary computing to design novel molecules for cancer treatment. To optimise drug efficacy, combat drug resistance, and reduce side effects in cancer therapy, we propose a two-birds-with-one-stone approach to design drugs that can target multiple drug targets. Below shows a compound (generated using the Lab's multi-target drug design method) that can bind two targets for breast cancer treatment.
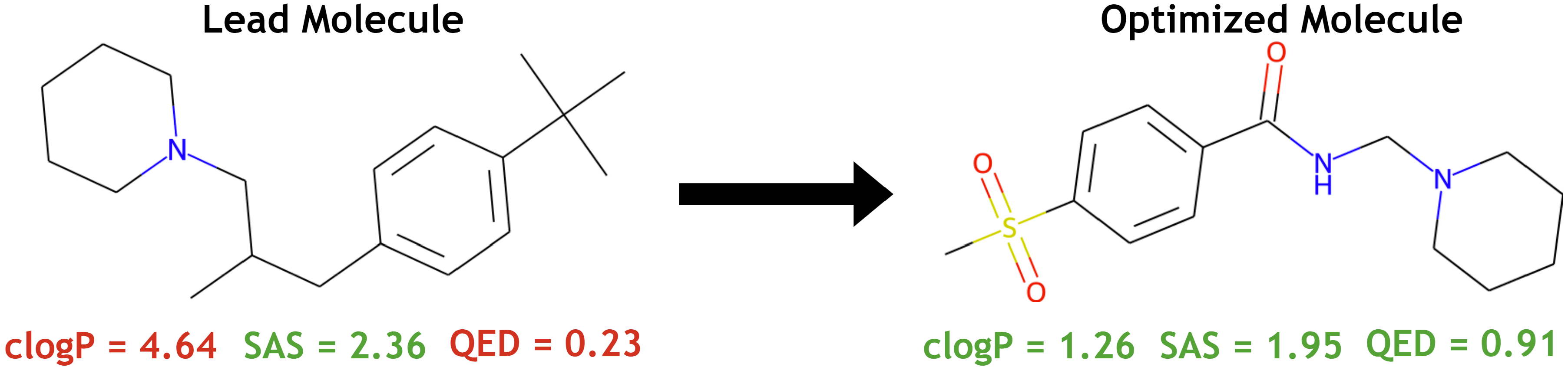
From the computing perspective, drug design is a multi-objective optimisation problem [1]. One particular task is lead optimisation which aims at modifying a given lead molecule of interest such that the desired properties can be improved and the unwanted properties minimised. Below demonstrates an example. The lead compound (left) has favourable SAS (synthetic accessibility score) property, but poor solubility in water (clogP) and drug-likeness (QED). After modification using BMDS' multi-objective reinforcement learning (MORL) approach, the new compound (right) has all properties favourable.
Biomedical Image Analysis
From images captured under microscopes to images taken in filed trips, image processing and analytics play an important role in scientific discovery. The BMDS Lab builds AI models for a variety of biomedical image analysis tasks, such as object segmentation and image classification. In collaboration with the 8Trees Inc., an environmental organisation in Font Hill Ontario, AI models are being developed to identify and track individual Massasauga rattlesnakes, an endangered species in South Ontario. Snake IDing is a challenging task due to their high-similar skin patterns and flexible body shape. From the two snake pictures below, are you able to tell whether they are from the same individual or two different ones? AI can tell.


Machine Learning
As an important pillar of AI, machine learning aims at building a parameterised model of our “world” of interest, learning its parameters using training examples and experiences, and then generalising its capability to unseen data. At the BMDS Lab, a variety of novel machine learning models have been devised to tackle specific tasks, such as deep feature selection (DFS) for cis-regulatory element prediction and characterisation, multi-class non-negative matrix factorisation for identifying shared pathways and tumor-specific pathways in pan-cancer analysis, and multi-modal generative models for data integration.
The BMDS group builds foundation models that can do multiple tasks and conversational AI models that can generate better responses given instructions. See below a demonstration.
Instruction: Write an e-mail to congratulate new Brock University admits and mention that you are excited about meeting all of them in person.
Text generated using BMDS' finetuned model:
Dear [Name],
Congratulations on being admitted to Brock University! I am thrilled to hear that you have been
accepted to
this prestigious institution and I am excited to meet you in person.
Brock University is known for its high-quality education, research, and innovation, and I am
sure
that you
will have a rewarding and enriching experience during your time here.
I would like to extend a warm welcome to you and wish you all the best in your studies. I am
looking
forward
to meeting you and getting to know you better.
Once again, congratulations on your admission and I hope to see you soon.
Best wishes,
[Your Name]
Text generated using the original model:
The Brock University Admissions Office will send you an e-mail with your Brock University
student
number and
instructions on how to access your Brock University student portal.
You will be able to access your Brock University student portal to check your Brock University
grades, Brock
University class schedule, Brock University student email, Brock University... [repeated
contents]
Looking Forward
At the BMDS Lab, we aim to push the boundaries of biomedical data science through impactful and interdisciplinary research. We actively seek new collaborations across academia, industry, and government sectors to translate our innovations into real-world solutions.
Selected Publications
See our publications page for a full list.
- A. Al-Jumaily, M. Mukaidaisi, A. Vu, A. Tchagang, Y. Li. Examining multi-objective deep reinforcement learning frameworks for molecular design. Biosystems, 232:104989, 2023.
- C. Andress, K. Kappel, M. E. Villena, M. Cupperlovic-Culf, H. Yan, Y. Li. DAPTEV: Deep aptamer evolutionary modelling for COVID-19 drug design. PLoS Computational Biology, 19(7):e1010774, 2023.
- M. Mukaidaisi, A. Vu, K. Grantham, A. Tchagang, Y. Li. Multi-objective drug design based on graph-fragment molecular representation and deep evolutionary learning. Frontiers in Pharmacology, 13:920747, 2022.
- K. Grantham, M. Mukaidaisi, H. K. Ooi, M. S. Ghaemi, A. Tchagang, Y. Li. Deep evolutionary learning for molecular design. IEEE Computational Intelligence Magazine, 17(2):14-28, 2022.
- Y. Li, Y. Pan, Z. Liu. Multi-class non-negative matrix factorization for comprehensive feature pattern discovery. IEEE Transactions on Neural Networks and Learning Systems, 30(2):615-629, 2019.
- Y. Li, F. Wu, A. Ngom. A review on machine learning principles for multi-view biological data integration. Briefings in Bioinformatics, 19(2):325-340, 2018.
- Y. Li, W. Shi, W. Wasserman. Genome-wide prediction of cis-regulatory regions using supervised deep learning methods. BMC Bioinformatics, 19:202, 2018.
